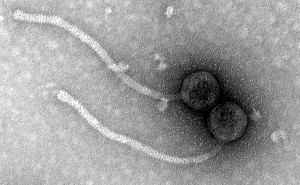
 We produce TEM images of bacteriophage for research groups and the SEA-PHAGES program. We provide several images per sample for morphological measurements with at least one “headshot” for a journal figure or the PhagesDB.
We produce TEM images of bacteriophage for research groups and the SEA-PHAGES program. We provide several images per sample for morphological measurements with at least one “headshot” for a journal figure or the PhagesDB.
We accept prepared grids and/or lysate. Imaging requests can typically be performed within a week of receipt. Samples can be imaged live via teleconference, which requires at least a two-week lead time for scheduling.
You will be registered in the Bookitlab core facility management system, where you will need to submit a KPIF:PHAGES Imaging service request when you are ready to ship your samples. Resulting images will be provided via a shared Google Drive and need to be downloaded within one week. We do not store data or samples.
Please acknowledge the KPIF in resulting publications and share them with us!